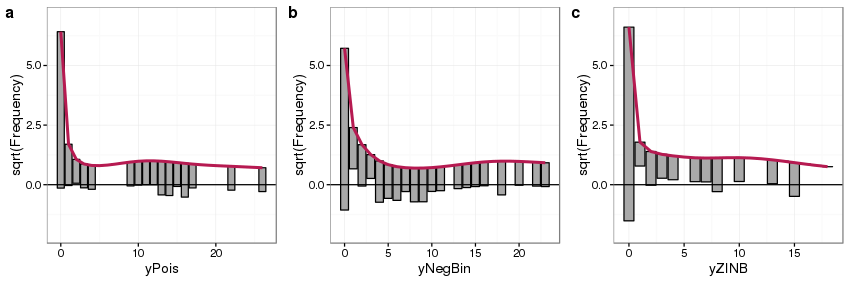
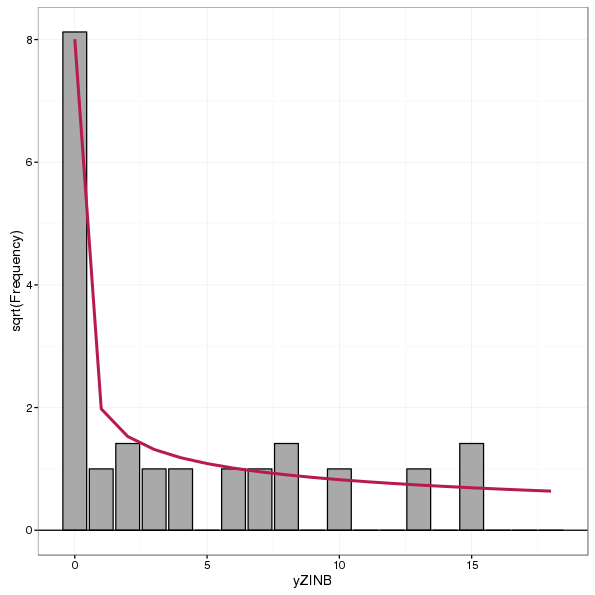
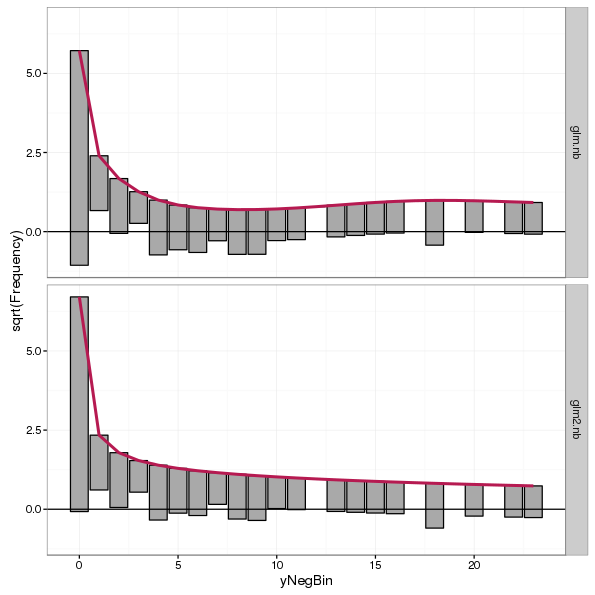
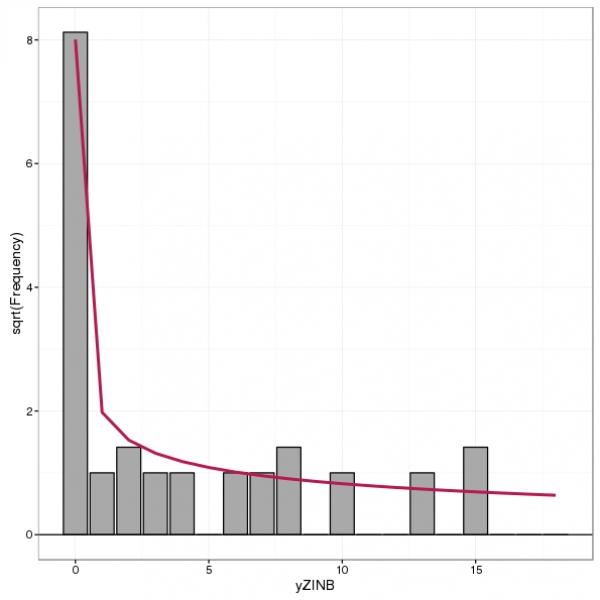
Query Time: 1.02 ms Query memory: 0.005 MB Memory before query: 4.295 MB Rows returned: 1
SELECT `session_id`
FROM `w0v41_session`
WHERE `session_id` = X'3033613731356636363436653933663433663261333765396630626238343531'
LIMIT 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_session | NULL | const | PRIMARY | PRIMARY | 194 | const | 1 | 100.00 | Using index |
| Status | Duration |
|---|
| starting | 0.29 ms |
| Executing hook on transaction | 0.02 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.24 ms |
| init | 0.01 ms |
| System lock | 0.02 ms |
| optimizing | 0.02 ms |
| statistics | 0.10 ms |
| preparing | 0.02 ms |
| executing | 0.02 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.02 ms |
| closing tables | 0.01 ms |
| freeing items | 0.09 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 9 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 8 | JDatabaseDriver->loadResult() | JROOT/libraries/src/Session/MetadataManager.php:74 |
| 7 | Joomla\CMS\Session\MetadataManager->createRecordIfNonExisting() | JROOT/libraries/src/Application/CMSApplication.php:154 |
| 6 | Joomla\CMS\Application\CMSApplication->checkSession() | JROOT/libraries/src/Application/CMSApplication.php:828 |
| 5 | Joomla\CMS\Application\CMSApplication->loadSession() | JROOT/libraries/src/Application/CMSApplication.php:136 |
| 4 | Joomla\CMS\Application\CMSApplication->__construct() | JROOT/libraries/src/Application/SiteApplication.php:66 |
| 3 | Joomla\CMS\Application\SiteApplication->__construct() | JROOT/libraries/src/Application/CMSApplication.php:386 |
| 2 | Joomla\CMS\Application\CMSApplication::getInstance() | JROOT/libraries/src/Factory.php:140 |
| 1 | Joomla\CMS\Factory::getApplication() | JROOT/index.php:46 |
Query Time: 0.59 ms After last query: 12.97 ms Query memory: 0.005 MB Memory before query: 4.883 MB Rows returned: 7
SELECT id, rules
FROM `w0v41_viewlevels`
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_viewlevels | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 7 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.15 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.02 ms |
| preparing | 0.02 ms |
| executing | 0.06 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 10 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1506 |
| 9 | JDatabaseDriver->loadAssocList() | JROOT/libraries/src/Access/Access.php:1063 |
| 8 | Joomla\CMS\Access\Access::getAuthorisedViewLevels() | JROOT/libraries/src/User/User.php:458 |
| 7 | Joomla\CMS\User\User->getAuthorisedViewLevels() | JROOT/libraries/src/Plugin/PluginHelper.php:318 |
| 6 | Joomla\CMS\Plugin\PluginHelper::load() | JROOT/libraries/src/Plugin/PluginHelper.php:87 |
| 5 | Joomla\CMS\Plugin\PluginHelper::getPlugin() | JROOT/libraries/src/Plugin/PluginHelper.php:129 |
| 4 | Joomla\CMS\Plugin\PluginHelper::isEnabled() | JROOT/libraries/src/Application/SiteApplication.php:604 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.70 ms After last query: 0.08 ms Query memory: 0.005 MB Memory before query: 4.896 MB Rows returned: 2
SELECT b.id
FROM w0v41_usergroups AS a
LEFT JOIN w0v41_usergroups AS b
ON b.lft <= a.lft
AND b.rgt >= a.rgt
WHERE a.id = 9
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | a | NULL | const | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| 1 | SIMPLE | b | NULL | range | idx_usergroup_nested_set_lookup | idx_usergroup_nested_set_lookup | 4 | NULL | 2 | 100.00 | Using where; Using index |
| Status | Duration |
|---|
| starting | 0.11 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.22 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.10 ms |
| preparing | 0.03 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 11 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1550 |
| 10 | JDatabaseDriver->loadColumn() | JROOT/libraries/src/Access/Access.php:980 |
| 9 | Joomla\CMS\Access\Access::getGroupsByUser() | JROOT/libraries/src/Access/Access.php:1095 |
| 8 | Joomla\CMS\Access\Access::getAuthorisedViewLevels() | JROOT/libraries/src/User/User.php:458 |
| 7 | Joomla\CMS\User\User->getAuthorisedViewLevels() | JROOT/libraries/src/Plugin/PluginHelper.php:318 |
| 6 | Joomla\CMS\Plugin\PluginHelper::load() | JROOT/libraries/src/Plugin/PluginHelper.php:87 |
| 5 | Joomla\CMS\Plugin\PluginHelper::getPlugin() | JROOT/libraries/src/Plugin/PluginHelper.php:129 |
| 4 | Joomla\CMS\Plugin\PluginHelper::isEnabled() | JROOT/libraries/src/Application/SiteApplication.php:604 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.75 ms After last query: 48.98 ms Query memory: 0.005 MB Memory before query: 7.595 MB Rows returned: 1
SELECT `name`
FROM `w0v41_extensions`
WHERE `type` = 'package'
AND `element` = 'pkg_eventgallery_full'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_extensions | NULL | ref | element_clientid,element_folder_clientid,extension | element_clientid | 402 | const | 1 | 10.00 | Using where |
| Status | Duration |
|---|
| starting | 0.14 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.15 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.10 ms |
| preparing | 0.02 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 10 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 9 | JDatabaseDriver->loadResult() | JROOT/administrator/components/com_eventgallery/version.php:24 |
| 8 | include_once JROOT/administrator/components/com_eventgallery/version.php | JROOT/plugins/system/picasaupdater/picasaupdater.php:32 |
| 7 | plgSystemPicasaupdater->__construct() | JROOT/libraries/src/Plugin/PluginHelper.php:280 |
| 6 | Joomla\CMS\Plugin\PluginHelper::import() | JROOT/libraries/src/Plugin/PluginHelper.php:182 |
| 5 | Joomla\CMS\Plugin\PluginHelper::importPlugin() | JROOT/libraries/src/Application/CMSApplication.php:667 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.80 ms After last query: 23.02 ms Query memory: 0.007 MB Memory before query: 8.286 MB Rows returned: 1
SELECT template
FROM w0v41_template_styles as s
WHERE s.client_id = 0
AND s.home = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | s | NULL | ref | idx_client_id,idx_client_id_home | idx_client_id | 1 | const | 6 | 12.50 | Using where |
| Status | Duration |
|---|
| starting | 0.14 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.21 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.06 ms |
| preparing | 0.02 ms |
| executing | 0.08 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 14 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 13 | JDatabaseDriver->loadResult() | JROOT/libraries/rokcommon/RokCommon/PlatformInfo/Joomla.php:31 |
| 12 | RokCommon_PlatformInfo_Joomla->getDefaultTemplate() | JROOT/libraries/rokcommon/RokCommon/PlatformInfo/Joomla.php:105 |
| 11 | RokCommon_PlatformInfo_Joomla->setPlatformParameters() | JROOT/libraries/rokcommon/RokCommon/Service.php:71 |
| 10 | RokCommon_Service::getContainer() | JROOT/libraries/rokcommon/include.php:38 |
| 9 | require_once JROOT/libraries/rokcommon/include.php | JROOT/plugins/system/rokcommon/rokcommon.php:95 |
| 8 | plgSystemRokCommon->loadCommonLib() | JROOT/plugins/system/rokcommon/rokcommon.php:53 |
| 7 | plgSystemRokCommon->__construct() | JROOT/libraries/src/Plugin/PluginHelper.php:280 |
| 6 | Joomla\CMS\Plugin\PluginHelper::import() | JROOT/libraries/src/Plugin/PluginHelper.php:182 |
| 5 | Joomla\CMS\Plugin\PluginHelper::importPlugin() | JROOT/libraries/src/Application/CMSApplication.php:667 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.43 ms After last query: 0.05 ms Query memory: 0.007 MB Memory before query: 8.294 MB Rows returned: 1
SELECT template
FROM w0v41_template_styles as s
WHERE s.client_id = 0
AND s.home = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | s | NULL | ref | idx_client_id,idx_client_id_home | idx_client_id | 1 | const | 6 | 12.50 | Using where |
| Status | Duration |
|---|
| starting | 0.09 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.02 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.05 ms |
| preparing | 0.02 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 14 | JDatabaseDriver->loadResult() | JROOT/libraries/rokcommon/RokCommon/PlatformInfo/Joomla.php:31 |
| 13 | RokCommon_PlatformInfo_Joomla->getDefaultTemplate() | JROOT/libraries/rokcommon/RokCommon/PlatformInfo/Joomla.php:67 |
| 12 | RokCommon_PlatformInfo_Joomla->getDefaultTemplatePath() | JROOT/libraries/rokcommon/RokCommon/PlatformInfo/Joomla.php:106 |
| 11 | RokCommon_PlatformInfo_Joomla->setPlatformParameters() | JROOT/libraries/rokcommon/RokCommon/Service.php:71 |
| 10 | RokCommon_Service::getContainer() | JROOT/libraries/rokcommon/include.php:38 |
| 9 | require_once JROOT/libraries/rokcommon/include.php | JROOT/plugins/system/rokcommon/rokcommon.php:95 |
| 8 | plgSystemRokCommon->loadCommonLib() | JROOT/plugins/system/rokcommon/rokcommon.php:53 |
| 7 | plgSystemRokCommon->__construct() | JROOT/libraries/src/Plugin/PluginHelper.php:280 |
| 6 | Joomla\CMS\Plugin\PluginHelper::import() | JROOT/libraries/src/Plugin/PluginHelper.php:182 |
| 5 | Joomla\CMS\Plugin\PluginHelper::importPlugin() | JROOT/libraries/src/Application/CMSApplication.php:667 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.62 ms After last query: 8.59 ms Query memory: 0.005 MB Memory before query: 8.540 MB Rows returned: 2
SELECT extension, file, type
FROM w0v41_rokcommon_configs
ORDER BY priority
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_rokcommon_configs | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 2 | 100.00 | Using filesort |
| Status | Duration |
|---|
| starting | 0.13 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.17 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.03 ms |
| preparing | 0.02 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 10 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 9 | JDatabaseDriver->loadObjectList() | JROOT/plugins/system/rokcommon/rokcommon.php:131 |
| 8 | plgSystemRokCommon->processRegisteredConfigs() | JROOT/plugins/system/rokcommon/rokcommon.php:75 |
| 7 | plgSystemRokCommon->__construct() | JROOT/libraries/src/Plugin/PluginHelper.php:280 |
| 6 | Joomla\CMS\Plugin\PluginHelper::import() | JROOT/libraries/src/Plugin/PluginHelper.php:182 |
| 5 | Joomla\CMS\Plugin\PluginHelper::importPlugin() | JROOT/libraries/src/Application/CMSApplication.php:667 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.29 ms After last query: 77.63 ms Query memory: 0.006 MB Memory before query: 12.417 MB Rows returned: 69
SELECT *
FROM w0v41_rsform_config
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_rsform_config | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 69 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.19 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.27 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.02 ms |
| preparing | 0.02 ms |
| executing | 0.45 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 13 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 12 | JDatabaseDriver->loadObjectList() | JROOT/administrator/components/com_rsform/helpers/config.php:50 |
| 11 | RSFormProConfig->load() | JROOT/administrator/components/com_rsform/helpers/config.php:17 |
| 10 | RSFormProConfig->__construct() | JROOT/administrator/components/com_rsform/helpers/config.php:102 |
| 9 | RSFormProConfig::getInstance() | JROOT/plugins/system/rsformdeletesubmissions/rsformdeletesubmissions.php:28 |
| 8 | plgSystemRsformdeletesubmissions->onAfterInitialise() | JROOT/libraries/joomla/event/event.php:70 |
| 7 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 6 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 5 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/CMSApplication.php:668 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 3.18 ms After last query: 4.31 ms Query memory: 0.007 MB Memory before query: 12.599 MB Rows returned: 20
SHOW FULL COLUMNS
FROM `w0v41_extensions`
EXPLAIN not possible on query: SHOW FULL COLUMNS FROM `w0v41_extensions`
| Status | Duration |
|---|
| starting | 0.17 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.07 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.94 ms |
| init | 0.02 ms |
| System lock | 0.02 ms |
| optimizing | 0.04 ms |
| statistics | 0.19 ms |
| preparing | 0.05 ms |
| Creating tmp table | 0.10 ms |
| executing | 0.13 ms |
| checking permissions | 0.11 ms |
| checking permissions | 0.06 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.02 ms |
| checking permissions | 0.02 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.06 ms |
| checking permissions | 0.06 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.12 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.03 ms |
| closing tables | 0.03 ms |
| freeing items | 0.14 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 14 | JDatabaseDriver->loadObjectList() | JROOT/libraries/joomla/database/driver/mysqli.php:448 |
| 13 | JDatabaseDriverMysqli->getTableColumns() | JROOT/libraries/src/Table/Table.php:261 |
| 12 | Joomla\CMS\Table\Table->getFields() | JROOT/libraries/src/Table/Table.php:180 |
| 11 | Joomla\CMS\Table\Table->__construct() | JROOT/libraries/src/Table/Extension.php:32 |
| 10 | Joomla\CMS\Table\Extension->__construct() | JROOT/libraries/src/Table/Table.php:328 |
| 9 | Joomla\CMS\Table\Table::getInstance() | JROOT/plugins/system/roksprocket/roksprocket.php:45 |
| 8 | plgSystemRokSprocket->onAfterInitialise() | JROOT/libraries/joomla/event/event.php:70 |
| 7 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 6 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 5 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/CMSApplication.php:668 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.65 ms After last query: 2.13 ms Query memory: 0.005 MB Memory before query: 12.716 MB Rows returned: 1
SELECT `extension_id`
FROM `w0v41_extensions`
WHERE type = 'component'
AND element = 'com_roksprocket'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_extensions | NULL | ref | element_clientid,element_folder_clientid,extension | extension | 484 | const,const | 1 | 100.00 | Using index |
| Status | Duration |
|---|
| starting | 0.14 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.03 ms |
| init | 0.01 ms |
| System lock | 0.03 ms |
| optimizing | 0.02 ms |
| statistics | 0.13 ms |
| preparing | 0.02 ms |
| executing | 0.04 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 11 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 10 | JDatabaseDriver->loadResult() | JROOT/libraries/src/Table/Extension.php:124 |
| 9 | Joomla\CMS\Table\Extension->find() | JROOT/plugins/system/roksprocket/roksprocket.php:47 |
| 8 | plgSystemRokSprocket->onAfterInitialise() | JROOT/libraries/joomla/event/event.php:70 |
| 7 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 6 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 5 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/CMSApplication.php:668 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.54 ms After last query: 0.08 ms Query memory: 0.005 MB Memory before query: 12.722 MB Rows returned: 1
SELECT *
FROM w0v41_extensions
WHERE `extension_id` = '10274'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_extensions | NULL | const | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.05 ms |
| preparing | 0.02 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 11 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1465 |
| 10 | JDatabaseDriver->loadAssoc() | JROOT/libraries/src/Table/Table.php:747 |
| 9 | Joomla\CMS\Table\Table->load() | JROOT/plugins/system/roksprocket/roksprocket.php:54 |
| 8 | plgSystemRokSprocket->onAfterInitialise() | JROOT/libraries/joomla/event/event.php:70 |
| 7 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 6 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 5 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/CMSApplication.php:668 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.53 ms After last query: 0.08 ms Query memory: 0.005 MB Memory before query: 12.727 MB Rows returned: 1
SELECT `extension_id`
FROM `w0v41_extensions`
WHERE type = 'module'
AND element = 'mod_roksprocket'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_extensions | NULL | ref | element_clientid,element_folder_clientid,extension | extension | 484 | const,const | 1 | 100.00 | Using index |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.03 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.08 ms |
| preparing | 0.02 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 11 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 10 | JDatabaseDriver->loadResult() | JROOT/libraries/src/Table/Extension.php:124 |
| 9 | Joomla\CMS\Table\Extension->find() | JROOT/plugins/system/roksprocket/roksprocket.php:70 |
| 8 | plgSystemRokSprocket->onAfterInitialise() | JROOT/libraries/joomla/event/event.php:70 |
| 7 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 6 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 5 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/CMSApplication.php:668 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.50 ms After last query: 0.06 ms Query memory: 0.005 MB Memory before query: 12.732 MB Rows returned: 1
SELECT *
FROM w0v41_extensions
WHERE `extension_id` = '10274'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_extensions | NULL | const | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.11 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.03 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.04 ms |
| preparing | 0.02 ms |
| executing | 0.04 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.09 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 11 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1465 |
| 10 | JDatabaseDriver->loadAssoc() | JROOT/libraries/src/Table/Table.php:747 |
| 9 | Joomla\CMS\Table\Table->load() | JROOT/plugins/system/roksprocket/roksprocket.php:76 |
| 8 | plgSystemRokSprocket->onAfterInitialise() | JROOT/libraries/joomla/event/event.php:70 |
| 7 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 6 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 5 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/CMSApplication.php:668 |
| 4 | Joomla\CMS\Application\CMSApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:686 |
| 3 | Joomla\CMS\Application\SiteApplication->initialiseApp() | JROOT/libraries/src/Application/SiteApplication.php:212 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.15 ms After last query: 182.20 ms Query memory: 0.006 MB Memory before query: 14.689 MB Rows returned: 0
SELECT id, alias, parent
FROM w0v41_k2_categories
WHERE published = 1
AND alias = 'rootograms,-a-new-way-to-assess-count-models'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_categories | NULL | ref | category,published,idx_category | category | 2 | const | 14 | 10.00 | Using where |
| Status | Duration |
|---|
| starting | 0.24 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.32 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.11 ms |
| preparing | 0.03 ms |
| executing | 0.14 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 13 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 12 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/router.php:493 |
| 11 | getCategoryProps() | JROOT/components/com_k2/router.php:313 |
| 10 | k2ParseRoute() | JROOT/libraries/src/Component/Router/RouterLegacy.php:105 |
| 9 | Joomla\CMS\Component\Router\RouterLegacy->parse() | JROOT/libraries/src/Router/SiteRouter.php:438 |
| 8 | Joomla\CMS\Router\SiteRouter->parseSefRoute() | JROOT/libraries/src/Router/Router.php:482 |
| 7 | Joomla\CMS\Router\Router->_parseSefRoute() | JROOT/libraries/src/Router/Router.php:227 |
| 6 | Joomla\CMS\Router\Router->parse() | JROOT/libraries/src/Router/SiteRouter.php:139 |
| 5 | Joomla\CMS\Router\SiteRouter->parse() | JROOT/libraries/src/Application/CMSApplication.php:1142 |
| 4 | Joomla\CMS\Application\CMSApplication->route() | JROOT/libraries/src/Application/SiteApplication.php:796 |
| 3 | Joomla\CMS\Application\SiteApplication->route() | JROOT/libraries/src/Application/SiteApplication.php:218 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.17 ms After last query: 0.05 ms Query memory: 0.006 MB Memory before query: 14.695 MB Rows returned: 1
SELECT id, alias
FROM w0v41_k2_items
WHERE published = 1
AND alias = 'rootograms,-a-new-way-to-assess-count-models'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_items | NULL | ref | idx_item | idx_item | 2 | const | 84 | 10.00 | Using where |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.17 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.08 ms |
| preparing | 0.02 ms |
| executing | 0.52 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 13 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 12 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/router.php:470 |
| 11 | getItemProps() | JROOT/components/com_k2/router.php:341 |
| 10 | k2ParseRoute() | JROOT/libraries/src/Component/Router/RouterLegacy.php:105 |
| 9 | Joomla\CMS\Component\Router\RouterLegacy->parse() | JROOT/libraries/src/Router/SiteRouter.php:438 |
| 8 | Joomla\CMS\Router\SiteRouter->parseSefRoute() | JROOT/libraries/src/Router/Router.php:482 |
| 7 | Joomla\CMS\Router\Router->_parseSefRoute() | JROOT/libraries/src/Router/Router.php:227 |
| 6 | Joomla\CMS\Router\Router->parse() | JROOT/libraries/src/Router/SiteRouter.php:139 |
| 5 | Joomla\CMS\Router\SiteRouter->parse() | JROOT/libraries/src/Application/CMSApplication.php:1142 |
| 4 | Joomla\CMS\Application\CMSApplication->route() | JROOT/libraries/src/Application/SiteApplication.php:796 |
| 3 | Joomla\CMS\Application\SiteApplication->route() | JROOT/libraries/src/Application/SiteApplication.php:218 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.12 ms After last query: 111.92 ms Query memory: 0.006 MB Memory before query: 16.950 MB
UPDATE `w0v41_extensions`
SET `params` = '{\"mediaversion\":\"3cb70a19691fdf174f01c352e8ac565c\"}'
WHERE `type` = 'library'
AND `element` = 'joomla'
EXPLAIN not possible on query: UPDATE `w0v41_extensions`
SET `params` = '{\"mediaversion\":\"3cb70a19691fdf174f01c352e8ac565c\"}'
WHERE `type` = 'library' AND `element` = 'joomla'
| Status | Duration |
|---|
| starting | 0.19 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.07 ms |
| init | 0.01 ms |
| System lock | 0.14 ms |
| updating | 0.13 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.32 ms |
| closing tables | 0.02 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 14 | JDatabaseDriverMysqli->execute() | JROOT/libraries/src/Helper/LibraryHelper.php:117 |
| 13 | Joomla\CMS\Helper\LibraryHelper::saveParams() | JROOT/libraries/src/Version.php:372 |
| 12 | Joomla\CMS\Version->setMediaVersion() | JROOT/libraries/src/Version.php:331 |
| 11 | Joomla\CMS\Version->getMediaVersion() | JROOT/libraries/src/Factory.php:778 |
| 10 | Joomla\CMS\Factory::createDocument() | JROOT/libraries/src/Factory.php:234 |
| 9 | Joomla\CMS\Factory::getDocument() | JROOT/plugins/system/k2/k2.php:374 |
| 8 | plgSystemK2->onAfterRoute() | JROOT/libraries/joomla/event/event.php:70 |
| 7 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 6 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 5 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/CMSApplication.php:1190 |
| 4 | Joomla\CMS\Application\CMSApplication->route() | JROOT/libraries/src/Application/SiteApplication.php:796 |
| 3 | Joomla\CMS\Application\SiteApplication->route() | JROOT/libraries/src/Application/SiteApplication.php:218 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 2.11 ms After last query: 25.69 ms Query memory: 0.007 MB Memory before query: 17.729 MB Rows returned: 60
SELECT `id`,`name`,`rules`,`parent_id`
FROM `w0v41_assets`
WHERE `name` IN ('root.1','com_actionlogs','com_acym','com_acymailing','com_admin','com_adminmenumanager','com_advportfoliopro','com_ajax','com_akeeba','com_associations','com_banners','com_bdthemes_shortcodes','com_cache','com_categories','com_checkin','com_cjlib','com_config','com_contact','com_content','com_contenthistory','com_cpanel','com_eventgallery','com_extplorer','com_fields','com_finder','com_installer','com_jce','com_jmap','com_joominapayments','com_joomlaupdate','com_k2','com_languages','com_login','com_mailto','com_media','com_menus','com_messages','com_modules','com_newsfeeds','com_plugins','com_postinstall','com_privacy','com_profiles','com_redirect','com_roksprocket','com_rsform','com_search','com_sigpro','com_smartcountdown3','com_speventum','com_splms','com_spmedical','com_sppagebuilder','com_spsimpleportfolio','com_tags','com_teamchart','com_templates','com_tinypayment','com_uniterevolution2','com_users','com_wrapper')
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_assets | NULL | range | idx_asset_name | idx_asset_name | 202 | NULL | 61 | 100.00 | Using index condition |
| Status | Duration |
|---|
| starting | 0.23 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.25 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.31 ms |
| preparing | 0.04 ms |
| executing | 0.95 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 13 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 12 | JDatabaseDriver->loadObjectList() | JROOT/libraries/src/Access/Access.php:429 |
| 11 | Joomla\CMS\Access\Access::preloadComponents() | JROOT/libraries/src/Access/Access.php:213 |
| 10 | Joomla\CMS\Access\Access::preload() | JROOT/libraries/src/Access/Access.php:531 |
| 9 | Joomla\CMS\Access\Access::getAssetRules() | JROOT/libraries/src/Access/Access.php:183 |
| 8 | Joomla\CMS\Access\Access::check() | JROOT/libraries/src/User/User.php:398 |
| 7 | Joomla\CMS\User\User->authorise() | JROOT/components/com_k2/k2.php:15 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.80 ms After last query: 19.26 ms Query memory: 0.007 MB Memory before query: 18.694 MB Rows returned: 13
SELECT id
FROM w0v41_k2_categories
WHERE published=1
AND trash=0
AND access IN(1,1,5)
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_categories | NULL | ref | category,published,access,trash,idx_category | trash | 2 | const | 13 | 100.00 | Using where |
| Status | Duration |
|---|
| starting | 0.16 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.16 ms |
| preparing | 0.03 ms |
| executing | 0.13 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1550 |
| 14 | JDatabaseDriver->loadColumn() | JROOT/components/com_k2/helpers/permissions.php:225 |
| 13 | K2HelperPermissions::canAddItem() | JROOT/components/com_k2/views/item/view.html.php:37 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.98 ms After last query: 0.09 ms Query memory: 0.007 MB Memory before query: 18.701 MB Rows returned: 1
SELECT *
FROM w0v41_k2_items
WHERE id=9
AND language IN ('en-GB', '*')
LIMIT 0, 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_items | NULL | const | PRIMARY,language | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.14 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.08 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.09 ms |
| preparing | 0.02 ms |
| executing | 0.39 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.03 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 14 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/models/item.php:33 |
| 13 | K2ModelItem->getData() | JROOT/components/com_k2/views/item/view.html.php:44 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 2.49 ms After last query: 1.41 ms Query memory: 0.009 MB Memory before query: 18.776 MB Rows returned: 14
SHOW FULL COLUMNS
FROM `w0v41_k2_categories`
EXPLAIN not possible on query: SHOW FULL COLUMNS FROM `w0v41_k2_categories`
| Status | Duration |
|---|
| starting | 0.15 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.06 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.86 ms |
| init | 0.02 ms |
| System lock | 0.02 ms |
| optimizing | 0.04 ms |
| statistics | 0.16 ms |
| preparing | 0.04 ms |
| Creating tmp table | 0.07 ms |
| executing | 0.11 ms |
| checking permissions | 0.10 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.02 ms |
| checking permissions | 0.02 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.09 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.02 ms |
| closing tables | 0.02 ms |
| freeing items | 0.13 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 19 | JDatabaseDriver->loadObjectList() | JROOT/libraries/joomla/database/driver/mysqli.php:448 |
| 18 | JDatabaseDriverMysqli->getTableColumns() | JROOT/libraries/src/Table/Table.php:261 |
| 17 | Joomla\CMS\Table\Table->getFields() | JROOT/libraries/src/Table/Table.php:180 |
| 16 | Joomla\CMS\Table\Table->__construct() | JROOT/administrator/components/com_k2/tables/k2category.php:34 |
| 15 | TableK2Category->__construct() | JROOT/libraries/src/Table/Table.php:328 |
| 14 | Joomla\CMS\Table\Table::getInstance() | JROOT/components/com_k2/models/item.php:64 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.53 ms After last query: 0.10 ms Query memory: 0.007 MB Memory before query: 18.804 MB Rows returned: 1
SELECT *
FROM w0v41_k2_categories
WHERE id = '3'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_categories | NULL | const | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.03 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.06 ms |
| preparing | 0.02 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 16 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1465 |
| 15 | JDatabaseDriver->loadAssoc() | JROOT/administrator/components/com_k2/tables/k2category.php:61 |
| 14 | TableK2Category->load() | JROOT/components/com_k2/models/item.php:65 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.52 ms After last query: 2.25 ms Query memory: 0.011 MB Memory before query: 18.846 MB Rows returned: 1
SELECT id, alias, parent
FROM w0v41_k2_categories
WHERE published = 1
AND id = 3
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_categories | NULL | const | PRIMARY,category,published,idx_category | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.14 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.03 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.05 ms |
| preparing | 0.02 ms |
| executing | 0.02 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.03 ms |
| # | Caller | File and line number |
|---|
| 25 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 24 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/router.php:493 |
| 23 | getCategoryProps() | JROOT/components/com_k2/router.php:501 |
| 22 | getCategoryPath() | JROOT/components/com_k2/router.php:205 |
| 21 | k2BuildRoute() | JROOT/libraries/src/Component/Router/RouterLegacy.php:69 |
| 20 | Joomla\CMS\Component\Router\RouterLegacy->build() | JROOT/libraries/src/Router/SiteRouter.php:532 |
| 19 | Joomla\CMS\Router\SiteRouter->buildSefRoute() | JROOT/libraries/src/Router/SiteRouter.php:502 |
| 18 | Joomla\CMS\Router\SiteRouter->_buildSefRoute() | JROOT/libraries/src/Router/Router.php:281 |
| 17 | Joomla\CMS\Router\Router->build() | JROOT/libraries/src/Router/SiteRouter.php:155 |
| 16 | Joomla\CMS\Router\SiteRouter->build() | JROOT/libraries/src/Router/Route.php:144 |
| 15 | Joomla\CMS\Router\Route::link() | JROOT/libraries/src/Router/Route.php:93 |
| 14 | Joomla\CMS\Router\Route::_() | JROOT/components/com_k2/models/item.php:68 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.85 ms After last query: 1.49 ms Query memory: 0.007 MB Memory before query: 18.911 MB Rows returned: 0
SELECT tag.*
FROM w0v41_k2_tags AS tag
JOIN w0v41_k2_tags_xref AS xref
ON tag.id = xref.tagID
WHERE tag.published = 1
AND xref.itemID = 9
ORDER BY xref.id ASC| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | xref | NULL | ref | tagID,itemID | itemID | 4 | const | 1 | 100.00 | NULL |
| 1 | SIMPLE | tag | NULL | eq_ref | PRIMARY,published | PRIMARY | 4 | h88876_SDAT.xref.tagID | 1 | 100.00 | Using where |
| Status | Duration |
|---|
| starting | 0.14 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.29 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.08 ms |
| preparing | 0.03 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 16 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 15 | JDatabaseDriver->loadObjectList() | JROOT/components/com_k2/models/item.php:1201 |
| 14 | K2ModelItem->getItemTags() | JROOT/components/com_k2/models/item.php:111 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.60 ms After last query: 0.25 ms Query memory: 0.007 MB Memory before query: 18.919 MB Rows returned: 0
SELECT *
FROM w0v41_k2_attachments
WHERE itemID=9
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_attachments | NULL | ref | itemID | itemID | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.14 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.05 ms |
| preparing | 0.03 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.09 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 16 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 15 | JDatabaseDriver->loadObjectList() | JROOT/components/com_k2/models/item.php:1429 |
| 14 | K2ModelItem->getItemAttachments() | JROOT/components/com_k2/models/item.php:163 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.79 ms After last query: 0.04 ms Query memory: 0.008 MB Memory before query: 18.927 MB Rows returned: 1
SELECT *
FROM w0v41_k2_rating
WHERE itemID = 9
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_rating | NULL | const | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.11 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 1.32 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.05 ms |
| preparing | 0.01 ms |
| executing | 0.02 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.04 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 17 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 16 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/models/item.php:914 |
| 15 | K2ModelItem->getRating() | JROOT/components/com_k2/models/item.php:958 |
| 14 | K2ModelItem->getVotesPercentage() | JROOT/components/com_k2/models/item.php:168 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 2.53 ms After last query: 1.50 ms Query memory: 0.011 MB Memory before query: 19.001 MB Rows returned: 17
SHOW FULL COLUMNS
FROM `w0v41_users`
EXPLAIN not possible on query: SHOW FULL COLUMNS FROM `w0v41_users`
| Status | Duration |
|---|
| starting | 0.15 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.06 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.78 ms |
| init | 0.02 ms |
| System lock | 0.01 ms |
| optimizing | 0.04 ms |
| statistics | 0.15 ms |
| preparing | 0.05 ms |
| Creating tmp table | 0.07 ms |
| executing | 0.12 ms |
| checking permissions | 0.09 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.02 ms |
| checking permissions | 0.02 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.09 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.02 ms |
| closing tables | 0.02 ms |
| freeing items | 0.14 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 25 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 24 | JDatabaseDriver->loadObjectList() | JROOT/libraries/joomla/database/driver/mysqli.php:448 |
| 23 | JDatabaseDriverMysqli->getTableColumns() | JROOT/libraries/src/Table/Table.php:261 |
| 22 | Joomla\CMS\Table\Table->getFields() | JROOT/libraries/src/Table/Table.php:180 |
| 21 | Joomla\CMS\Table\Table->__construct() | JROOT/libraries/src/Table/User.php:41 |
| 20 | Joomla\CMS\Table\User->__construct() | JROOT/libraries/src/Table/Table.php:328 |
| 19 | Joomla\CMS\Table\Table::getInstance() | JROOT/libraries/src/User/User.php:603 |
| 18 | Joomla\CMS\User\User::getTable() | JROOT/libraries/src/User/User.php:877 |
| 17 | Joomla\CMS\User\User->load() | JROOT/libraries/src/User/User.php:248 |
| 16 | Joomla\CMS\User\User->__construct() | JROOT/libraries/src/User/User.php:301 |
| 15 | Joomla\CMS\User\User::getInstance() | JROOT/libraries/src/Factory.php:266 |
| 14 | Joomla\CMS\Factory::getUser() | JROOT/components/com_k2/models/item.php:208 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.63 ms After last query: 0.11 ms Query memory: 0.009 MB Memory before query: 19.039 MB Rows returned: 1
SELECT *
FROM `w0v41_users`
WHERE `id` = 111
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_users | NULL | const | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.16 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.06 ms |
| preparing | 0.02 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.09 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1465 |
| 19 | JDatabaseDriver->loadAssoc() | JROOT/libraries/src/Table/User.php:87 |
| 18 | Joomla\CMS\Table\User->load() | JROOT/libraries/src/User/User.php:880 |
| 17 | Joomla\CMS\User\User->load() | JROOT/libraries/src/User/User.php:248 |
| 16 | Joomla\CMS\User\User->__construct() | JROOT/libraries/src/User/User.php:301 |
| 15 | Joomla\CMS\User\User::getInstance() | JROOT/libraries/src/Factory.php:266 |
| 14 | Joomla\CMS\Factory::getUser() | JROOT/components/com_k2/models/item.php:208 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.74 ms After last query: 4.28 ms Query memory: 0.009 MB Memory before query: 19.856 MB Rows returned: 3
SELECT `g`.`id`,`g`.`title`
FROM `w0v41_usergroups` AS g
INNER JOIN `w0v41_user_usergroup_map` AS m
ON m.group_id = g.id
WHERE `m`.`user_id` = 111
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | m | NULL | ref | PRIMARY | PRIMARY | 4 | const | 3 | 100.00 | Using index |
| 1 | SIMPLE | g | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.m.group_id | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.14 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.17 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.08 ms |
| preparing | 0.02 ms |
| executing | 0.06 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1506 |
| 19 | JDatabaseDriver->loadAssocList() | JROOT/libraries/src/Table/User.php:112 |
| 18 | Joomla\CMS\Table\User->load() | JROOT/libraries/src/User/User.php:880 |
| 17 | Joomla\CMS\User\User->load() | JROOT/libraries/src/User/User.php:248 |
| 16 | Joomla\CMS\User\User->__construct() | JROOT/libraries/src/User/User.php:301 |
| 15 | Joomla\CMS\User\User::getInstance() | JROOT/libraries/src/Factory.php:266 |
| 14 | Joomla\CMS\Factory::getUser() | JROOT/components/com_k2/models/item.php:208 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.70 ms After last query: 1.02 ms Query memory: 0.008 MB Memory before query: 19.864 MB Rows returned: 1
SELECT id, gender, description, image, url, `
group`, plugins
FROM w0v41_k2_users
WHERE userID=111
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_users | NULL | ref | userID | userID | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.15 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.17 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.05 ms |
| preparing | 0.02 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 17 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 16 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/models/item.php:1681 |
| 15 | K2ModelItem->getUserProfile() | JROOT/components/com_k2/helpers/utilities.php:45 |
| 14 | K2HelperUtilities::getAvatar() | JROOT/components/com_k2/models/item.php:211 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:83 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.65 ms After last query: 13.48 ms Query memory: 0.009 MB Memory before query: 20.363 MB Rows returned: 1
SELECT enabled
FROM w0v41_extensions
WHERE element = 'com_sppagebuilder'
AND type = 'component'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_extensions | NULL | ref | element_clientid,element_folder_clientid,extension | element_clientid | 402 | const | 1 | 10.00 | Using where |
| Status | Duration |
|---|
| starting | 0.15 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.11 ms |
| preparing | 0.03 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 19 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 18 | JDatabaseDriver->loadResult() | JROOT/plugins/content/sppagebuilder/sppagebuilder.php:172 |
| 17 | PlgContentSppagebuilder->isSppagebuilderEnabled() | JROOT/plugins/content/sppagebuilder/sppagebuilder.php:36 |
| 16 | PlgContentSppagebuilder->__construct() | JROOT/libraries/src/Plugin/PluginHelper.php:280 |
| 15 | Joomla\CMS\Plugin\PluginHelper::import() | JROOT/libraries/src/Plugin/PluginHelper.php:182 |
| 14 | Joomla\CMS\Plugin\PluginHelper::importPlugin() | JROOT/components/com_k2/models/item.php:488 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:87 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.98 ms After last query: 13.69 ms Query memory: 0.010 MB Memory before query: 20.851 MB Rows returned: 0
SELECT DISTINCT a.id, a.title, a.name, a.checked_out, a.checked_out_time, a.note, a.state, a.access, a.created_time, a.created_user_id, a.ordering, a.language, a.fieldparams, a.params, a.type, a.default_value, a.context, a.group_id, a.label, a.description, a.required,l.title AS language_title, l.image AS language_image,uc.name AS editor,ag.title AS access_level,ua.name AS author_name,g.title AS group_title, g.access as group_access, g.state AS group_state, g.note as group_note
FROM w0v41_fields AS a
LEFT JOIN `w0v41_languages` AS l
ON l.lang_code = a.language
LEFT JOIN w0v41_users AS uc
ON uc.id=a.checked_out
LEFT JOIN w0v41_viewlevels AS ag
ON ag.id = a.access
LEFT JOIN w0v41_users AS ua
ON ua.id = a.created_user_id
LEFT JOIN w0v41_fields_groups AS g
ON g.id = a.group_id
WHERE a.context = 'com_k2.item-media'
AND a.access IN (1,1,5)
AND (a.group_id = 0 OR g.access IN (1,1,5))
AND a.state = 1
AND (a.group_id = 0 OR g.state = 1)
ORDER BY a.ordering ASC
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | a | NULL | ref | idx_state,idx_access,idx_context | idx_state | 1 | const | 1 | 100.00 | Using where; Using temporary; Using filesort |
| 1 | SIMPLE | l | NULL | eq_ref | idx_langcode | idx_langcode | 28 | h88876_SDAT.a.language | 1 | 100.00 | Using index condition |
| 1 | SIMPLE | uc | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.checked_out | 1 | 100.00 | NULL |
| 1 | SIMPLE | ag | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.access | 1 | 100.00 | Using where |
| 1 | SIMPLE | ua | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.created_user_id | 1 | 100.00 | Using where |
| 1 | SIMPLE | g | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.group_id | 1 | 100.00 | Using where |
| Status | Duration |
|---|
| starting | 0.32 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.68 ms |
| init | 0.02 ms |
| System lock | 0.02 ms |
| optimizing | 0.04 ms |
| statistics | 0.26 ms |
| preparing | 0.05 ms |
| Creating tmp table | 0.10 ms |
| executing | 0.15 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.03 ms |
| closing tables | 0.03 ms |
| freeing items | 0.12 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 22 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 21 | JDatabaseDriver->loadObjectList() | JROOT/libraries/src/MVC/Model/BaseDatabaseModel.php:322 |
| 20 | Joomla\CMS\MVC\Model\BaseDatabaseModel->_getList() | JROOT/administrator/components/com_fields/models/fields.php:333 |
| 19 | FieldsModelFields->_getList() | JROOT/libraries/src/MVC/Model/ListModel.php:194 |
| 18 | Joomla\CMS\MVC\Model\ListModel->getItems() | JROOT/administrator/components/com_fields/helpers/fields.php:136 |
| 17 | FieldsHelper::getFields() | JROOT/plugins/system/fields/fields.php:495 |
| 16 | PlgSystemFields->onContentPrepare() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:596 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:87 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.80 ms After last query: 25.24 ms Query memory: 0.010 MB Memory before query: 21.609 MB Rows returned: 0
SELECT DISTINCT a.id, a.title, a.name, a.checked_out, a.checked_out_time, a.note, a.state, a.access, a.created_time, a.created_user_id, a.ordering, a.language, a.fieldparams, a.params, a.type, a.default_value, a.context, a.group_id, a.label, a.description, a.required,l.title AS language_title, l.image AS language_image,uc.name AS editor,ag.title AS access_level,ua.name AS author_name,g.title AS group_title, g.access as group_access, g.state AS group_state, g.note as group_note
FROM w0v41_fields AS a
LEFT JOIN `w0v41_languages` AS l
ON l.lang_code = a.language
LEFT JOIN w0v41_users AS uc
ON uc.id=a.checked_out
LEFT JOIN w0v41_viewlevels AS ag
ON ag.id = a.access
LEFT JOIN w0v41_users AS ua
ON ua.id = a.created_user_id
LEFT JOIN w0v41_fields_groups AS g
ON g.id = a.group_id
LEFT JOIN `w0v41_fields_categories` AS fc
ON fc.field_id = a.id
WHERE a.context = 'com_k2.item'
AND (fc.category_id IS NULL OR fc.category_id IN (3,0))
AND a.access IN (1,1,5)
AND (a.group_id = 0 OR g.access IN (1,1,5))
AND a.state = 1
AND (a.group_id = 0 OR g.state = 1)
AND a.language in ('*','en-GB')
ORDER BY a.ordering ASC
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | a | NULL | ref | idx_state,idx_access,idx_context,idx_language | idx_state | 1 | const | 1 | 100.00 | Using where; Using temporary; Using filesort |
| 1 | SIMPLE | l | NULL | eq_ref | idx_langcode | idx_langcode | 28 | h88876_SDAT.a.language | 1 | 100.00 | Using index condition |
| 1 | SIMPLE | uc | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.checked_out | 1 | 100.00 | NULL |
| 1 | SIMPLE | ag | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.access | 1 | 100.00 | Using where |
| 1 | SIMPLE | ua | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.created_user_id | 1 | 100.00 | Using where |
| 1 | SIMPLE | g | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.group_id | 1 | 100.00 | Using where |
| 1 | SIMPLE | fc | NULL | ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.id | 1 | 100.00 | Using where; Using index; Distinct |
| Status | Duration |
|---|
| starting | 0.41 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.02 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.32 ms |
| init | 0.02 ms |
| System lock | 0.02 ms |
| optimizing | 0.05 ms |
| statistics | 0.32 ms |
| preparing | 0.06 ms |
| Creating tmp table | 0.12 ms |
| executing | 0.12 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.03 ms |
| closing tables | 0.02 ms |
| freeing items | 0.12 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 22 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 21 | JDatabaseDriver->loadObjectList() | JROOT/libraries/src/MVC/Model/BaseDatabaseModel.php:322 |
| 20 | Joomla\CMS\MVC\Model\BaseDatabaseModel->_getList() | JROOT/administrator/components/com_fields/models/fields.php:333 |
| 19 | FieldsModelFields->_getList() | JROOT/libraries/src/MVC/Model/ListModel.php:194 |
| 18 | Joomla\CMS\MVC\Model\ListModel->getItems() | JROOT/administrator/components/com_fields/helpers/fields.php:136 |
| 17 | FieldsHelper::getFields() | JROOT/plugins/system/fields/fields.php:495 |
| 16 | PlgSystemFields->onContentPrepare() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:667 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:87 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.70 ms After last query: 11.41 ms Query memory: 0.010 MB Memory before query: 22.091 MB Rows returned: 1
SELECT enabled
FROM w0v41_extensions
WHERE element = 'com_sppagebuilder'
AND type = 'component'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_extensions | NULL | ref | element_clientid,element_folder_clientid,extension | element_clientid | 402 | const | 1 | 10.00 | Using where |
| Status | Duration |
|---|
| starting | 0.17 ms |
| Executing hook on transaction | 0.02 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.12 ms |
| preparing | 0.03 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 19 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 18 | JDatabaseDriver->loadResult() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:99 |
| 17 | plgK2Sppagebuilder->isSppagebuilderEnabled() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:84 |
| 16 | plgK2Sppagebuilder->onK2PrepareContent() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:792 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:87 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 2.54 ms After last query: 0.41 ms Query memory: 0.009 MB Memory before query: 22.107 MB Rows returned: 0
SELECT *
FROM `w0v41_sppagebuilder`
WHERE `extension` = 'com_k2'
AND `extension_view` = 'item'
AND `view_id` = '9'
AND `active` = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_sppagebuilder | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 131 | 0.76 | Using where |
| Status | Duration |
|---|
| starting | 0.15 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.21 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.02 ms |
| preparing | 0.03 ms |
| executing | 1.79 ms |
| end | 0.03 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.13 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 19 | JDatabaseDriver->loadObject() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:291 |
| 18 | SppagebuilderHelper::getPageContent() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:248 |
| 17 | SppagebuilderHelper::onIntegrationPrepareContent() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:87 |
| 16 | plgK2Sppagebuilder->onK2PrepareContent() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:792 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:87 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 2.02 ms After last query: 0.07 ms Query memory: 0.009 MB Memory before query: 22.116 MB Rows returned: 0
SELECT *
FROM `w0v41_sppagebuilder`
WHERE `extension` = 'com_k2'
AND `extension_view` = 'item'
AND `view_id` = '9'
AND `active` = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_sppagebuilder | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 131 | 0.76 | Using where |
| Status | Duration |
|---|
| starting | 0.15 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.06 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.03 ms |
| statistics | 0.03 ms |
| preparing | 0.03 ms |
| executing | 1.43 ms |
| end | 0.02 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.02 ms |
| closing tables | 0.02 ms |
| freeing items | 0.12 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 19 | JDatabaseDriver->loadObject() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:291 |
| 18 | SppagebuilderHelper::getPageContent() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:248 |
| 17 | SppagebuilderHelper::onIntegrationPrepareContent() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:89 |
| 16 | plgK2Sppagebuilder->onK2PrepareContent() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:792 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:87 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 3.72 ms After last query: 1.31 ms Query memory: 0.009 MB Memory before query: 22.167 MB Rows returned: 36
SHOW FULL COLUMNS
FROM `w0v41_k2_items`
EXPLAIN not possible on query: SHOW FULL COLUMNS FROM `w0v41_k2_items`
| Status | Duration |
|---|
| starting | 0.19 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.09 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.92 ms |
| init | 0.02 ms |
| System lock | 0.02 ms |
| optimizing | 0.04 ms |
| statistics | 0.18 ms |
| preparing | 0.05 ms |
| Creating tmp table | 0.08 ms |
| executing | 0.14 ms |
| checking permissions | 0.11 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.02 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.03 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.05 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.04 ms |
| checking permissions | 0.13 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.03 ms |
| closing tables | 0.03 ms |
| freeing items | 0.15 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 19 | JDatabaseDriver->loadObjectList() | JROOT/libraries/joomla/database/driver/mysqli.php:448 |
| 18 | JDatabaseDriverMysqli->getTableColumns() | JROOT/libraries/src/Table/Table.php:261 |
| 17 | Joomla\CMS\Table\Table->getFields() | JROOT/libraries/src/Table/Table.php:180 |
| 16 | Joomla\CMS\Table\Table->__construct() | JROOT/administrator/components/com_k2/tables/k2item.php:56 |
| 15 | TableK2Item->__construct() | JROOT/libraries/src/Table/Table.php:328 |
| 14 | Joomla\CMS\Table\Table::getInstance() | JROOT/components/com_k2/models/item.php:839 |
| 13 | K2ModelItem->hit() | JROOT/components/com_k2/views/item/view.html.php:146 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.18 ms After last query: 0.14 ms Query memory: 0.007 MB Memory before query: 22.227 MB
UPDATE w0v41_k2_items
SET `hits` = (`hits` + 1)
WHERE `id` = '9'
EXPLAIN not possible on query: UPDATE w0v41_k2_items
SET `hits` = (`hits` + 1)
WHERE `id` = '9'
| Status | Duration |
|---|
| starting | 0.14 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.05 ms |
| init | 0.01 ms |
| System lock | 0.10 ms |
| updating | 0.19 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.48 ms |
| closing tables | 0.02 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/src/Table/Table.php:1264 |
| 14 | Joomla\CMS\Table\Table->hit() | JROOT/components/com_k2/models/item.php:840 |
| 13 | K2ModelItem->hit() | JROOT/components/com_k2/views/item/view.html.php:146 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.78 ms After last query: 0.12 ms Query memory: 0.007 MB Memory before query: 22.231 MB Rows returned: 0
SELECT *
FROM w0v41_k2_comments
WHERE itemID=9
AND published=1
ORDER BY commentDate DESC
LIMIT 0, 10
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_comments | NULL | ref | itemID,published,latestComments,countComments | itemID | 4 | const | 1 | 100.00 | Using where; Using filesort |
| Status | Duration |
|---|
| starting | 0.14 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.20 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.09 ms |
| preparing | 0.04 ms |
| executing | 0.04 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 14 | JDatabaseDriver->loadObjectList() | JROOT/components/com_k2/models/item.php:1450 |
| 13 | K2ModelItem->getItemComments() | JROOT/components/com_k2/views/item/view.html.php:227 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 4.48 ms After last query: 2.19 ms Query memory: 0.007 MB Memory before query: 22.321 MB Rows returned: 5
SELECT i.*, c.alias AS categoryalias
FROM w0v41_k2_items AS i
LEFT JOIN w0v41_k2_categories c
ON c.id = i.catid
WHERE i.id != 9
AND i.published = 1
AND (i.publish_up = '0000-00-00 00:00:00' OR i.publish_up <= '2025-08-29 19:22:52')
AND (i.publish_down = '0000-00-00 00:00:00' OR i.publish_down >= '2025-08-29 19:22:52')
AND i.access IN(1,1,5)
AND i.language IN('en-GB', '*')
AND i.trash = 0
AND i.created_by = 111
AND i.created_by_alias=''
AND c.published = 1
AND c.access IN(1,1,5)
AND c.language IN('en-GB', '*')
AND c.trash = 0
ORDER BY i.created DESC
LIMIT 0, 5| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | i | NULL | range | PRIMARY,catid,created_by,language,idx_item | PRIMARY | 4 | NULL | 42 | 1.39 | Using where; Using filesort |
| 1 | SIMPLE | c | NULL | eq_ref | PRIMARY,category,published,access,trash,language,idx_category | PRIMARY | 4 | h88876_SDAT.i.catid | 1 | 57.14 | Using where |
| Status | Duration |
|---|
| starting | 0.22 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.11 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.06 ms |
| statistics | 0.81 ms |
| preparing | 0.07 ms |
| executing | 2.90 ms |
| end | 0.02 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.02 ms |
| closing tables | 0.02 ms |
| freeing items | 0.12 ms |
| cleaning up | 0.03 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 14 | JDatabaseDriver->loadObjectList() | JROOT/components/com_k2/models/itemlist.php:567 |
| 13 | K2ModelItemlist->getAuthorLatest() | JROOT/components/com_k2/views/item/view.html.php:266 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.05 ms After last query: 1.50 ms Query memory: 0.007 MB Memory before query: 22.521 MB Rows returned: 1
SELECT *
FROM w0v41_k2_items
WHERE id < 9
AND catid = 3
AND published = 1
AND trash = 0
AND (publish_up = '0000-00-00 00:00:00' OR publish_up <= '2025-08-29 19:22:52')
AND (publish_down = '0000-00-00 00:00:00' OR publish_down >= '2025-08-29 19:22:52')
AND access IN (1,1,5)
AND language IN ('en-GB', '*')
ORDER BY id DESC
LIMIT 0, 1| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_items | NULL | index_merge | PRIMARY,catid,language,idx_item | catid,PRIMARY | 8,4 | NULL | 1 | 5.00 | Using intersect(catid,PRIMARY); Using where; Using filesort |
| Status | Duration |
|---|
| starting | 0.20 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.08 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.05 ms |
| statistics | 0.18 ms |
| preparing | 0.05 ms |
| executing | 0.17 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.03 ms |
| closing tables | 0.02 ms |
| freeing items | 0.12 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 14 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/models/item.php:1604 |
| 13 | K2ModelItem->getPreviousItem() | JROOT/components/com_k2/views/item/view.html.php:290 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.73 ms After last query: 1.57 ms Query memory: 0.007 MB Memory before query: 22.598 MB Rows returned: 3
SELECT tag.*
FROM w0v41_k2_tags AS tag
JOIN w0v41_k2_tags_xref AS xref
ON tag.id = xref.tagID
WHERE tag.published = 1
AND xref.itemID = 1
ORDER BY xref.id ASC| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | xref | NULL | ref | tagID,itemID | itemID | 4 | const | 3 | 100.00 | NULL |
| 1 | SIMPLE | tag | NULL | eq_ref | PRIMARY,published | PRIMARY | 4 | h88876_SDAT.xref.tagID | 1 | 100.00 | Using where |
| Status | Duration |
|---|
| starting | 0.16 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.05 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.09 ms |
| preparing | 0.05 ms |
| executing | 0.08 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 16 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 15 | JDatabaseDriver->loadObjectList() | JROOT/components/com_k2/models/item.php:1201 |
| 14 | K2ModelItem->getItemTags() | JROOT/components/com_k2/models/item.php:111 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:292 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.55 ms After last query: 0.50 ms Query memory: 0.007 MB Memory before query: 22.612 MB Rows returned: 0
SELECT *
FROM w0v41_k2_attachments
WHERE itemID=1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_attachments | NULL | ref | itemID | itemID | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.13 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.05 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.06 ms |
| preparing | 0.02 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 16 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 15 | JDatabaseDriver->loadObjectList() | JROOT/components/com_k2/models/item.php:1429 |
| 14 | K2ModelItem->getItemAttachments() | JROOT/components/com_k2/models/item.php:163 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:292 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.51 ms After last query: 0.04 ms Query memory: 0.008 MB Memory before query: 22.620 MB Rows returned: 1
SELECT *
FROM w0v41_k2_rating
WHERE itemID = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_rating | NULL | const | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.05 ms |
| preparing | 0.02 ms |
| executing | 0.02 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 17 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 16 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/models/item.php:914 |
| 15 | K2ModelItem->getRating() | JROOT/components/com_k2/models/item.php:958 |
| 14 | K2ModelItem->getVotesPercentage() | JROOT/components/com_k2/models/item.php:168 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:292 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.54 ms After last query: 0.17 ms Query memory: 0.009 MB Memory before query: 22.633 MB Rows returned: 1
SELECT *
FROM `w0v41_users`
WHERE `id` = 110
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_users | NULL | const | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.11 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.07 ms |
| preparing | 0.02 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1465 |
| 19 | JDatabaseDriver->loadAssoc() | JROOT/libraries/src/Table/User.php:87 |
| 18 | Joomla\CMS\Table\User->load() | JROOT/libraries/src/User/User.php:880 |
| 17 | Joomla\CMS\User\User->load() | JROOT/libraries/src/User/User.php:248 |
| 16 | Joomla\CMS\User\User->__construct() | JROOT/libraries/src/User/User.php:301 |
| 15 | Joomla\CMS\User\User::getInstance() | JROOT/libraries/src/Factory.php:266 |
| 14 | Joomla\CMS\Factory::getUser() | JROOT/components/com_k2/models/item.php:208 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:292 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.58 ms After last query: 0.11 ms Query memory: 0.009 MB Memory before query: 22.646 MB Rows returned: 1
SELECT `g`.`id`,`g`.`title`
FROM `w0v41_usergroups` AS g
INNER JOIN `w0v41_user_usergroup_map` AS m
ON m.group_id = g.id
WHERE `m`.`user_id` = 110
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | m | NULL | ref | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | Using index |
| 1 | SIMPLE | g | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.m.group_id | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.13 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.08 ms |
| preparing | 0.02 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1506 |
| 19 | JDatabaseDriver->loadAssocList() | JROOT/libraries/src/Table/User.php:112 |
| 18 | Joomla\CMS\Table\User->load() | JROOT/libraries/src/User/User.php:880 |
| 17 | Joomla\CMS\User\User->load() | JROOT/libraries/src/User/User.php:248 |
| 16 | Joomla\CMS\User\User->__construct() | JROOT/libraries/src/User/User.php:301 |
| 15 | Joomla\CMS\User\User::getInstance() | JROOT/libraries/src/Factory.php:266 |
| 14 | Joomla\CMS\Factory::getUser() | JROOT/components/com_k2/models/item.php:208 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:292 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.51 ms After last query: 0.99 ms Query memory: 0.008 MB Memory before query: 22.652 MB Rows returned: 1
SELECT id, gender, description, image, url, `
group`, plugins
FROM w0v41_k2_users
WHERE userID=110
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_users | NULL | ref | userID | userID | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.13 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.03 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.05 ms |
| preparing | 0.02 ms |
| executing | 0.04 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 17 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 16 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/models/item.php:1681 |
| 15 | K2ModelItem->getUserProfile() | JROOT/components/com_k2/helpers/utilities.php:45 |
| 14 | K2HelperUtilities::getAvatar() | JROOT/components/com_k2/models/item.php:211 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:292 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.26 ms After last query: 0.70 ms Query memory: 0.010 MB Memory before query: 22.641 MB Rows returned: 0
SELECT DISTINCT a.id, a.title, a.name, a.checked_out, a.checked_out_time, a.note, a.state, a.access, a.created_time, a.created_user_id, a.ordering, a.language, a.fieldparams, a.params, a.type, a.default_value, a.context, a.group_id, a.label, a.description, a.required,l.title AS language_title, l.image AS language_image,uc.name AS editor,ag.title AS access_level,ua.name AS author_name,g.title AS group_title, g.access as group_access, g.state AS group_state, g.note as group_note
FROM w0v41_fields AS a
LEFT JOIN `w0v41_languages` AS l
ON l.lang_code = a.language
LEFT JOIN w0v41_users AS uc
ON uc.id=a.checked_out
LEFT JOIN w0v41_viewlevels AS ag
ON ag.id = a.access
LEFT JOIN w0v41_users AS ua
ON ua.id = a.created_user_id
LEFT JOIN w0v41_fields_groups AS g
ON g.id = a.group_id
WHERE a.context = 'com_k2.item-media'
AND a.access IN (1,1,5)
AND (a.group_id = 0 OR g.access IN (1,1,5))
AND a.state = 1
AND (a.group_id = 0 OR g.state = 1)
AND a.language in ('*','en-GB')
ORDER BY a.ordering ASC
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | a | NULL | ref | idx_state,idx_access,idx_context,idx_language | idx_state | 1 | const | 1 | 100.00 | Using where; Using temporary; Using filesort |
| 1 | SIMPLE | l | NULL | eq_ref | idx_langcode | idx_langcode | 28 | h88876_SDAT.a.language | 1 | 100.00 | Using index condition |
| 1 | SIMPLE | uc | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.checked_out | 1 | 100.00 | NULL |
| 1 | SIMPLE | ag | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.access | 1 | 100.00 | Using where |
| 1 | SIMPLE | ua | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.created_user_id | 1 | 100.00 | Using where |
| 1 | SIMPLE | g | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a.group_id | 1 | 100.00 | Using where |
| Status | Duration |
|---|
| starting | 0.25 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.13 ms |
| init | 0.03 ms |
| System lock | 0.02 ms |
| optimizing | 0.04 ms |
| statistics | 0.26 ms |
| preparing | 0.04 ms |
| Creating tmp table | 0.07 ms |
| executing | 0.10 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.02 ms |
| closing tables | 0.03 ms |
| freeing items | 0.10 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 22 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 21 | JDatabaseDriver->loadObjectList() | JROOT/libraries/src/MVC/Model/BaseDatabaseModel.php:322 |
| 20 | Joomla\CMS\MVC\Model\BaseDatabaseModel->_getList() | JROOT/administrator/components/com_fields/models/fields.php:333 |
| 19 | FieldsModelFields->_getList() | JROOT/libraries/src/MVC/Model/ListModel.php:194 |
| 18 | Joomla\CMS\MVC\Model\ListModel->getItems() | JROOT/administrator/components/com_fields/helpers/fields.php:136 |
| 17 | FieldsHelper::getFields() | JROOT/plugins/system/fields/fields.php:495 |
| 16 | PlgSystemFields->onContentPrepare() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:596 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:293 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.68 ms After last query: 3.61 ms Query memory: 0.009 MB Memory before query: 22.696 MB Rows returned: 1
SELECT enabled
FROM w0v41_extensions
WHERE element = 'com_sppagebuilder'
AND type = 'component'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_extensions | NULL | ref | element_clientid,element_folder_clientid,extension | element_clientid | 402 | const | 1 | 10.00 | Using where |
| Status | Duration |
|---|
| starting | 0.15 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.11 ms |
| preparing | 0.03 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.12 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 19 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 18 | JDatabaseDriver->loadResult() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:99 |
| 17 | plgK2Sppagebuilder->isSppagebuilderEnabled() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:84 |
| 16 | plgK2Sppagebuilder->onK2PrepareContent() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:792 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:293 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 2.28 ms After last query: 0.08 ms Query memory: 0.009 MB Memory before query: 22.707 MB Rows returned: 0
SELECT *
FROM `w0v41_sppagebuilder`
WHERE `extension` = 'com_k2'
AND `extension_view` = 'item'
AND `view_id` = '1'
AND `active` = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_sppagebuilder | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 131 | 0.76 | Using where |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.06 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.02 ms |
| preparing | 0.03 ms |
| executing | 1.74 ms |
| end | 0.02 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.12 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 19 | JDatabaseDriver->loadObject() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:291 |
| 18 | SppagebuilderHelper::getPageContent() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:248 |
| 17 | SppagebuilderHelper::onIntegrationPrepareContent() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:87 |
| 16 | plgK2Sppagebuilder->onK2PrepareContent() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:792 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:293 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.89 ms After last query: 0.07 ms Query memory: 0.009 MB Memory before query: 22.716 MB Rows returned: 0
SELECT *
FROM `w0v41_sppagebuilder`
WHERE `extension` = 'com_k2'
AND `extension_view` = 'item'
AND `view_id` = '1'
AND `active` = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_sppagebuilder | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 131 | 0.76 | Using where |
| Status | Duration |
|---|
| starting | 0.13 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.06 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.03 ms |
| preparing | 0.03 ms |
| executing | 1.33 ms |
| end | 0.02 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 19 | JDatabaseDriver->loadObject() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:291 |
| 18 | SppagebuilderHelper::getPageContent() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:248 |
| 17 | SppagebuilderHelper::onIntegrationPrepareContent() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:89 |
| 16 | plgK2Sppagebuilder->onK2PrepareContent() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:792 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:293 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.06 ms After last query: 0.29 ms Query memory: 0.007 MB Memory before query: 22.730 MB Rows returned: 1
SELECT *
FROM w0v41_k2_items
WHERE id > 9
AND catid = 3
AND published = 1
AND trash = 0
AND (publish_up = '0000-00-00 00:00:00' OR publish_up <= '2025-08-29 19:22:52')
AND (publish_down = '0000-00-00 00:00:00' OR publish_down >= '2025-08-29 19:22:52')
AND access IN (1,1,5)
AND language IN ('en-GB', '*')
ORDER BY id ASC
LIMIT 0, 1| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_items | NULL | range | PRIMARY,catid,language,idx_item | PRIMARY | 4 | NULL | 36 | 1.39 | Using where |
| Status | Duration |
|---|
| starting | 0.18 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.09 ms |
| init | 0.02 ms |
| System lock | 0.01 ms |
| optimizing | 0.05 ms |
| statistics | 0.21 ms |
| preparing | 0.04 ms |
| executing | 0.16 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.12 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 14 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/models/item.php:1663 |
| 13 | K2ModelItem->getNextItem() | JROOT/components/com_k2/views/item/view.html.php:331 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.73 ms After last query: 1.46 ms Query memory: 0.007 MB Memory before query: 22.806 MB Rows returned: 2
SELECT tag.*
FROM w0v41_k2_tags AS tag
JOIN w0v41_k2_tags_xref AS xref
ON tag.id = xref.tagID
WHERE tag.published = 1
AND xref.itemID = 11
ORDER BY xref.id ASC| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | xref | NULL | ref | tagID,itemID | itemID | 4 | const | 2 | 100.00 | NULL |
| 1 | SIMPLE | tag | NULL | eq_ref | PRIMARY,published | PRIMARY | 4 | h88876_SDAT.xref.tagID | 1 | 100.00 | Using where |
| Status | Duration |
|---|
| starting | 0.17 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.06 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.09 ms |
| preparing | 0.03 ms |
| executing | 0.08 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 16 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 15 | JDatabaseDriver->loadObjectList() | JROOT/components/com_k2/models/item.php:1201 |
| 14 | K2ModelItem->getItemTags() | JROOT/components/com_k2/models/item.php:111 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:333 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.50 ms After last query: 0.37 ms Query memory: 0.007 MB Memory before query: 22.818 MB Rows returned: 0
SELECT *
FROM w0v41_k2_attachments
WHERE itemID=11
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_k2_attachments | NULL | ref | itemID | itemID | 4 | const | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.11 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.03 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.05 ms |
| preparing | 0.02 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 16 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 15 | JDatabaseDriver->loadObjectList() | JROOT/components/com_k2/models/item.php:1429 |
| 14 | K2ModelItem->getItemAttachments() | JROOT/components/com_k2/models/item.php:163 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:333 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.41 ms After last query: 0.04 ms Query memory: 0.008 MB Memory before query: 22.826 MB Rows returned: 0
SELECT *
FROM w0v41_k2_rating
WHERE itemID = 11
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | NULL | NULL | NULL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | NULL | NULL | no matching row in const table |
| Status | Duration |
|---|
| starting | 0.10 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.04 ms |
| executing | 0.02 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 17 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 16 | JDatabaseDriver->loadObject() | JROOT/components/com_k2/models/item.php:914 |
| 15 | K2ModelItem->getRating() | JROOT/components/com_k2/models/item.php:958 |
| 14 | K2ModelItem->getVotesPercentage() | JROOT/components/com_k2/models/item.php:168 |
| 13 | K2ModelItem->prepareItem() | JROOT/components/com_k2/views/item/view.html.php:333 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.67 ms After last query: 4.49 ms Query memory: 0.009 MB Memory before query: 22.822 MB Rows returned: 1
SELECT enabled
FROM w0v41_extensions
WHERE element = 'com_sppagebuilder'
AND type = 'component'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_extensions | NULL | ref | element_clientid,element_folder_clientid,extension | element_clientid | 402 | const | 1 | 10.00 | Using where |
| Status | Duration |
|---|
| starting | 0.15 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.04 ms |
| init | 0.01 ms |
| System lock | 0.03 ms |
| optimizing | 0.02 ms |
| statistics | 0.11 ms |
| preparing | 0.03 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 19 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1740 |
| 18 | JDatabaseDriver->loadResult() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:99 |
| 17 | plgK2Sppagebuilder->isSppagebuilderEnabled() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:84 |
| 16 | plgK2Sppagebuilder->onK2PrepareContent() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:792 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:334 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.95 ms After last query: 0.07 ms Query memory: 0.009 MB Memory before query: 22.833 MB Rows returned: 0
SELECT *
FROM `w0v41_sppagebuilder`
WHERE `extension` = 'com_k2'
AND `extension_view` = 'item'
AND `view_id` = '11'
AND `active` = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_sppagebuilder | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 131 | 0.76 | Using where |
| Status | Duration |
|---|
| starting | 0.13 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.06 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.03 ms |
| preparing | 0.03 ms |
| executing | 1.41 ms |
| end | 0.02 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 19 | JDatabaseDriver->loadObject() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:291 |
| 18 | SppagebuilderHelper::getPageContent() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:248 |
| 17 | SppagebuilderHelper::onIntegrationPrepareContent() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:87 |
| 16 | plgK2Sppagebuilder->onK2PrepareContent() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:792 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:334 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.89 ms After last query: 0.06 ms Query memory: 0.009 MB Memory before query: 22.843 MB Rows returned: 0
SELECT *
FROM `w0v41_sppagebuilder`
WHERE `extension` = 'com_k2'
AND `extension_view` = 'item'
AND `view_id` = '11'
AND `active` = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_sppagebuilder | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 131 | 0.76 | Using where |
| Status | Duration |
|---|
| starting | 0.13 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.06 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.03 ms |
| statistics | 0.03 ms |
| preparing | 0.03 ms |
| executing | 1.34 ms |
| end | 0.02 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.02 ms |
| freeing items | 0.12 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 20 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 19 | JDatabaseDriver->loadObject() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:291 |
| 18 | SppagebuilderHelper::getPageContent() | JROOT/administrator/components/com_sppagebuilder/helpers/sppagebuilder.php:248 |
| 17 | SppagebuilderHelper::onIntegrationPrepareContent() | JROOT/plugins/k2/sppagebuilder/sppagebuilder.php:89 |
| 16 | plgK2Sppagebuilder->onK2PrepareContent() | JROOT/libraries/joomla/event/event.php:70 |
| 15 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 14 | JEventDispatcher->trigger() | JROOT/components/com_k2/models/item.php:792 |
| 13 | K2ModelItem->execPlugins() | JROOT/components/com_k2/views/item/view.html.php:334 |
| 12 | K2ViewItem->display() | JROOT/libraries/src/Cache/Controller/ViewController.php:102 |
| 11 | Joomla\CMS\Cache\Controller\ViewController->get() | JROOT/libraries/src/MVC/Controller/BaseController.php:655 |
| 10 | Joomla\CMS\MVC\Controller\BaseController->display() | JROOT/components/com_k2/controllers/controller.php:20 |
| 9 | K2Controller->display() | JROOT/components/com_k2/controllers/item.php:78 |
| 8 | K2ControllerItem->display() | JROOT/libraries/src/MVC/Controller/BaseController.php:702 |
| 7 | Joomla\CMS\MVC\Controller\BaseController->execute() | JROOT/components/com_k2/k2.php:57 |
| 6 | require_once JROOT/components/com_k2/k2.php | JROOT/libraries/src/Component/ComponentHelper.php:402 |
| 5 | Joomla\CMS\Component\ComponentHelper::executeComponent() | JROOT/libraries/src/Component/ComponentHelper.php:377 |
| 4 | Joomla\CMS\Component\ComponentHelper::renderComponent() | JROOT/libraries/src/Application/SiteApplication.php:194 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.27 ms After last query: 10.50 ms Query memory: 0.004 MB Memory before query: 23.010 MB Rows returned: 0
SELECT folder.*
FROM `w0v41_eventgallery_folder` AS folder
LEFT JOIN `w0v41_eventgallery_file` AS file
ON folder.folder = file.folder
and file.published=1
and file.ismainimage=0
WHERE file.file IS NULL
AND (folder.foldertypeid=1 OR folder.foldertypeid=2 OR folder.foldertypeid=4)
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | folder | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 4 | 57.81 | Using where |
| 1 | SIMPLE | file | NULL | ref | file,index_folder | file | 377 | h88876_SDAT.folder.folder | 38 | 10.00 | Using where; Not exists |
| Status | Duration |
|---|
| starting | 0.31 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.02 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.39 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.03 ms |
| statistics | 0.06 ms |
| preparing | 0.05 ms |
| executing | 0.10 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.02 ms |
| closing tables | 0.02 ms |
| freeing items | 0.11 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 9 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 8 | JDatabaseDriver->loadObjectList() | JROOT/plugins/system/picasaupdater/picasaupdater.php:80 |
| 7 | plgSystemPicasaupdater->onAfterDispatch() | JROOT/libraries/joomla/event/event.php:70 |
| 6 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 5 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 4 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/SiteApplication.php:199 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 1.31 ms After last query: 2.88 ms Query memory: 0.005 MB Memory before query: 23.075 MB Rows returned: 3
SELECT language,id
FROM `w0v41_menu`
WHERE home = 1
AND published = 1
AND client_id = 0
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_menu | NULL | ref | idx_client_id_parent_id_alias_language | idx_client_id_parent_id_alias_language | 1 | const | 216 | 1.00 | Using where |
| Status | Duration |
|---|
| starting | 0.13 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.14 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.07 ms |
| preparing | 0.02 ms |
| executing | 0.71 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.09 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 10 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 9 | JDatabaseDriver->loadObjectList() | JROOT/libraries/src/Language/Multilanguage.php:107 |
| 8 | Joomla\CMS\Language\Multilanguage::getSiteHomePages() | JROOT/plugins/system/languagefilter/languagefilter.php:751 |
| 7 | PlgSystemLanguageFilter->onAfterDispatch() | JROOT/libraries/joomla/event/event.php:70 |
| 6 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 5 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 4 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/SiteApplication.php:199 |
| 3 | Joomla\CMS\Application\SiteApplication->dispatch() | JROOT/libraries/src/Application/SiteApplication.php:233 |
| 2 | Joomla\CMS\Application\SiteApplication->doExecute() | JROOT/libraries/src/Application/CMSApplication.php:225 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 2.28 ms After last query: 123.91 ms Query memory: 0.007 MB Memory before query: 25.201 MB Rows returned: 159
SELECT *
FROM w0v41_acymailing_config
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_acymailing_config | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 159 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.23 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.24 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.02 ms |
| preparing | 0.03 ms |
| executing | 0.73 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.02 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 14 | JDatabaseDriver->loadObjectList() | JROOT/administrator/components/com_acymailing/classes/cpanel.php:17 |
| 13 | cpanelClass->load() | JROOT/administrator/components/com_acymailing/helpers/helper.php:452 |
| 12 | acymailing_config() | JROOT/administrator/components/com_acymailing/helpers/helper.php:1110 |
| 11 | include_once JROOT/administrator/components/com_acymailing/helpers/helper.php | JROOT/modules/mod_acymailing/mod_acymailing.php:12 |
| 10 | include JROOT/modules/mod_acymailing/mod_acymailing.php | JROOT/libraries/src/Helper/ModuleHelper.php:200 |
| 9 | Joomla\CMS\Helper\ModuleHelper::renderModule() | JROOT/libraries/src/Document/Renderer/Html/ModuleRenderer.php:98 |
| 8 | Joomla\CMS\Document\Renderer\Html\ModuleRenderer->render() | JROOT/libraries/src/Document/Renderer/Html/ModulesRenderer.php:47 |
| 7 | Joomla\CMS\Document\Renderer\Html\ModulesRenderer->render() | JROOT/libraries/src/Document/HtmlDocument.php:511 |
| 6 | Joomla\CMS\Document\HtmlDocument->getBuffer() | JROOT/libraries/src/Document/HtmlDocument.php:803 |
| 5 | Joomla\CMS\Document\HtmlDocument->_renderTemplate() | JROOT/libraries/src/Document/HtmlDocument.php:577 |
| 4 | Joomla\CMS\Document\HtmlDocument->render() | JROOT/libraries/src/Application/CMSApplication.php:1112 |
| 3 | Joomla\CMS\Application\CMSApplication->render() | JROOT/libraries/src/Application/SiteApplication.php:778 |
| 2 | Joomla\CMS\Application\SiteApplication->render() | JROOT/libraries/src/Application/CMSApplication.php:231 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.64 ms After last query: 2.71 ms Query memory: 0.006 MB Memory before query: 25.494 MB Rows returned: 3
SELECT *
FROM w0v41_acymailing_list
WHERE type = 'list'
ORDER BY ordering ASC
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_acymailing_list | NULL | ref | typeorderingindex,typeuseridindex | typeorderingindex | 1 | const | 2 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.11 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.16 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.07 ms |
| preparing | 0.02 ms |
| executing | 0.06 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 13 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 12 | JDatabaseDriver->loadObjectList() | JROOT/administrator/components/com_acymailing/classes/list.php:29 |
| 11 | listClass->getLists() | JROOT/modules/mod_acymailing/mod_acymailing.php:121 |
| 10 | include JROOT/modules/mod_acymailing/mod_acymailing.php | JROOT/libraries/src/Helper/ModuleHelper.php:200 |
| 9 | Joomla\CMS\Helper\ModuleHelper::renderModule() | JROOT/libraries/src/Document/Renderer/Html/ModuleRenderer.php:98 |
| 8 | Joomla\CMS\Document\Renderer\Html\ModuleRenderer->render() | JROOT/libraries/src/Document/Renderer/Html/ModulesRenderer.php:47 |
| 7 | Joomla\CMS\Document\Renderer\Html\ModulesRenderer->render() | JROOT/libraries/src/Document/HtmlDocument.php:511 |
| 6 | Joomla\CMS\Document\HtmlDocument->getBuffer() | JROOT/libraries/src/Document/HtmlDocument.php:803 |
| 5 | Joomla\CMS\Document\HtmlDocument->_renderTemplate() | JROOT/libraries/src/Document/HtmlDocument.php:577 |
| 4 | Joomla\CMS\Document\HtmlDocument->render() | JROOT/libraries/src/Application/CMSApplication.php:1112 |
| 3 | Joomla\CMS\Application\CMSApplication->render() | JROOT/libraries/src/Application/SiteApplication.php:778 |
| 2 | Joomla\CMS\Application\SiteApplication->render() | JROOT/libraries/src/Application/CMSApplication.php:231 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.63 ms After last query: 22.05 ms Query memory: 0.006 MB Memory before query: 27.165 MB Rows returned: 3
SELECT *
FROM `w0v41_acymailing_fields` as a
WHERE a.`published` = 1
AND (a.access = 'all' OR a.access LIKE ('%,9,%'))
ORDER BY a.`ordering` ASC
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | a | NULL | ref | orderingindex | orderingindex | 1 | const | 2 | 55.56 | Using where |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.15 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.04 ms |
| preparing | 0.02 ms |
| executing | 0.06 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 13 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 12 | JDatabaseDriver->loadObjectList() | JROOT/administrator/components/com_acymailing/classes/fields.php:91 |
| 11 | fieldsClass->getFields() | JROOT/modules/mod_acymailing/mod_acymailing.php:207 |
| 10 | include JROOT/modules/mod_acymailing/mod_acymailing.php | JROOT/libraries/src/Helper/ModuleHelper.php:200 |
| 9 | Joomla\CMS\Helper\ModuleHelper::renderModule() | JROOT/libraries/src/Document/Renderer/Html/ModuleRenderer.php:98 |
| 8 | Joomla\CMS\Document\Renderer\Html\ModuleRenderer->render() | JROOT/libraries/src/Document/Renderer/Html/ModulesRenderer.php:47 |
| 7 | Joomla\CMS\Document\Renderer\Html\ModulesRenderer->render() | JROOT/libraries/src/Document/HtmlDocument.php:511 |
| 6 | Joomla\CMS\Document\HtmlDocument->getBuffer() | JROOT/libraries/src/Document/HtmlDocument.php:803 |
| 5 | Joomla\CMS\Document\HtmlDocument->_renderTemplate() | JROOT/libraries/src/Document/HtmlDocument.php:577 |
| 4 | Joomla\CMS\Document\HtmlDocument->render() | JROOT/libraries/src/Application/CMSApplication.php:1112 |
| 3 | Joomla\CMS\Application\CMSApplication->render() | JROOT/libraries/src/Application/SiteApplication.php:778 |
| 2 | Joomla\CMS\Application\SiteApplication->render() | JROOT/libraries/src/Application/CMSApplication.php:231 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.37 ms After last query: 0.06 ms Query memory: 0.006 MB Memory before query: 27.181 MB Rows returned: 3
SELECT *
FROM `w0v41_acymailing_fields`
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_acymailing_fields | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 3 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.08 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.03 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.01 ms |
| preparing | 0.01 ms |
| executing | 0.04 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.07 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 13 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 12 | JDatabaseDriver->loadObjectList() | JROOT/administrator/components/com_acymailing/classes/fields.php:104 |
| 11 | fieldsClass->getFields() | JROOT/modules/mod_acymailing/mod_acymailing.php:207 |
| 10 | include JROOT/modules/mod_acymailing/mod_acymailing.php | JROOT/libraries/src/Helper/ModuleHelper.php:200 |
| 9 | Joomla\CMS\Helper\ModuleHelper::renderModule() | JROOT/libraries/src/Document/Renderer/Html/ModuleRenderer.php:98 |
| 8 | Joomla\CMS\Document\Renderer\Html\ModuleRenderer->render() | JROOT/libraries/src/Document/Renderer/Html/ModulesRenderer.php:47 |
| 7 | Joomla\CMS\Document\Renderer\Html\ModulesRenderer->render() | JROOT/libraries/src/Document/HtmlDocument.php:511 |
| 6 | Joomla\CMS\Document\HtmlDocument->getBuffer() | JROOT/libraries/src/Document/HtmlDocument.php:803 |
| 5 | Joomla\CMS\Document\HtmlDocument->_renderTemplate() | JROOT/libraries/src/Document/HtmlDocument.php:577 |
| 4 | Joomla\CMS\Document\HtmlDocument->render() | JROOT/libraries/src/Application/CMSApplication.php:1112 |
| 3 | Joomla\CMS\Application\CMSApplication->render() | JROOT/libraries/src/Application/SiteApplication.php:778 |
| 2 | Joomla\CMS\Application\SiteApplication->render() | JROOT/libraries/src/Application/CMSApplication.php:231 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.95 ms After last query: 13.63 ms Query memory: 0.007 MB Memory before query: 27.268 MB Rows returned: 2
SELECT `c2`.`language`,`c2`.`id`
FROM `w0v41_menu` AS `c`
INNER JOIN `w0v41_associations` AS `a`
ON a.id = c.`id`
AND a.context='com_menus.item'
INNER JOIN `w0v41_associations` AS `a2`
ON `a`.`key` = `a2`.`key`
INNER JOIN `w0v41_menu` AS `c2`
ON a2.id = c2.`id`
WHERE c.id = 197
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | c | NULL | const | PRIMARY | PRIMARY | 4 | const | 1 | 100.00 | Using index |
| 1 | SIMPLE | a | NULL | const | PRIMARY,idx_key | PRIMARY | 206 | const,const | 1 | 100.00 | NULL |
| 1 | SIMPLE | a2 | NULL | ref | idx_key | idx_key | 128 | const | 2 | 100.00 | Using index |
| 1 | SIMPLE | c2 | NULL | eq_ref | PRIMARY | PRIMARY | 4 | h88876_SDAT.a2.id | 1 | 100.00 | NULL |
| Status | Duration |
|---|
| starting | 0.12 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.36 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.15 ms |
| preparing | 0.02 ms |
| executing | 0.05 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 15 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1701 |
| 14 | JDatabaseDriver->loadObjectList() | JROOT/libraries/src/Language/Associations.php:115 |
| 13 | Joomla\CMS\Language\Associations::getAssociations() | JROOT/administrator/components/com_menus/helpers/menus.php:321 |
| 12 | MenusHelper::getAssociations() | JROOT/modules/mod_languages/helper.php:58 |
| 11 | ModLanguagesHelper::getList() | JROOT/modules/mod_languages/mod_languages.php:17 |
| 10 | include JROOT/modules/mod_languages/mod_languages.php | JROOT/libraries/src/Helper/ModuleHelper.php:200 |
| 9 | Joomla\CMS\Helper\ModuleHelper::renderModule() | JROOT/libraries/src/Document/Renderer/Html/ModuleRenderer.php:98 |
| 8 | Joomla\CMS\Document\Renderer\Html\ModuleRenderer->render() | JROOT/libraries/src/Document/Renderer/Html/ModulesRenderer.php:47 |
| 7 | Joomla\CMS\Document\Renderer\Html\ModulesRenderer->render() | JROOT/libraries/src/Document/HtmlDocument.php:511 |
| 6 | Joomla\CMS\Document\HtmlDocument->getBuffer() | JROOT/libraries/src/Document/HtmlDocument.php:803 |
| 5 | Joomla\CMS\Document\HtmlDocument->_renderTemplate() | JROOT/libraries/src/Document/HtmlDocument.php:577 |
| 4 | Joomla\CMS\Document\HtmlDocument->render() | JROOT/libraries/src/Application/CMSApplication.php:1112 |
| 3 | Joomla\CMS\Application\CMSApplication->render() | JROOT/libraries/src/Application/SiteApplication.php:778 |
| 2 | Joomla\CMS\Application\SiteApplication->render() | JROOT/libraries/src/Application/CMSApplication.php:231 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.56 ms After last query: 7.07 ms Query memory: 0.012 MB Memory before query: 27.344 MB Rows returned: 0
SELECT *
FROM w0v41_jmap_metainfo
WHERE `linkurl` = 'https://sdat.ir/en/sdat-blog/rootograms,-a-new-way-to-assess-count-models'
AND `published` = 1
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_jmap_metainfo | NULL | ref | published | published | 1 | const | 1 | 100.00 | Using where |
| Status | Duration |
|---|
| starting | 0.09 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.14 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.02 ms |
| statistics | 0.05 ms |
| preparing | 0.02 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.08 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 14 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 13 | JDatabaseDriver->loadObject() | JROOT/plugins/system/jmap/jmap.php:429 |
| 12 | plgSystemJMap->onBeforeCompileHead() | JROOT/libraries/joomla/event/event.php:70 |
| 11 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 10 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 9 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Document/Renderer/Html/HeadRenderer.php:67 |
| 8 | Joomla\CMS\Document\Renderer\Html\HeadRenderer->fetchHead() | JROOT/libraries/src/Document/Renderer/Html/HeadRenderer.php:38 |
| 7 | Joomla\CMS\Document\Renderer\Html\HeadRenderer->render() | JROOT/libraries/src/Document/HtmlDocument.php:511 |
| 6 | Joomla\CMS\Document\HtmlDocument->getBuffer() | JROOT/libraries/src/Document/HtmlDocument.php:803 |
| 5 | Joomla\CMS\Document\HtmlDocument->_renderTemplate() | JROOT/libraries/src/Document/HtmlDocument.php:577 |
| 4 | Joomla\CMS\Document\HtmlDocument->render() | JROOT/libraries/src/Application/CMSApplication.php:1112 |
| 3 | Joomla\CMS\Application\CMSApplication->render() | JROOT/libraries/src/Application/SiteApplication.php:778 |
| 2 | Joomla\CMS\Application\SiteApplication->render() | JROOT/libraries/src/Application/CMSApplication.php:231 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.46 ms After last query: 0.04 ms Query memory: 0.006 MB Memory before query: 27.356 MB Rows returned: 0
SELECT *
FROM w0v41_jmap_canonicals
WHERE `linkurl` = 'https://sdat.ir/en/sdat-blog/rootograms,-a-new-way-to-assess-count-models'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_jmap_canonicals | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 1 | 100.00 | Using where |
| Status | Duration |
|---|
| starting | 0.08 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.14 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.01 ms |
| preparing | 0.02 ms |
| executing | 0.02 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.07 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 14 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 13 | JDatabaseDriver->loadObject() | JROOT/plugins/system/jmap/jmap.php:508 |
| 12 | plgSystemJMap->onBeforeCompileHead() | JROOT/libraries/joomla/event/event.php:70 |
| 11 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 10 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 9 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Document/Renderer/Html/HeadRenderer.php:67 |
| 8 | Joomla\CMS\Document\Renderer\Html\HeadRenderer->fetchHead() | JROOT/libraries/src/Document/Renderer/Html/HeadRenderer.php:38 |
| 7 | Joomla\CMS\Document\Renderer\Html\HeadRenderer->render() | JROOT/libraries/src/Document/HtmlDocument.php:511 |
| 6 | Joomla\CMS\Document\HtmlDocument->getBuffer() | JROOT/libraries/src/Document/HtmlDocument.php:803 |
| 5 | Joomla\CMS\Document\HtmlDocument->_renderTemplate() | JROOT/libraries/src/Document/HtmlDocument.php:577 |
| 4 | Joomla\CMS\Document\HtmlDocument->render() | JROOT/libraries/src/Application/CMSApplication.php:1112 |
| 3 | Joomla\CMS\Application\CMSApplication->render() | JROOT/libraries/src/Application/SiteApplication.php:778 |
| 2 | Joomla\CMS\Application\SiteApplication->render() | JROOT/libraries/src/Application/CMSApplication.php:231 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
Query Time: 0.46 ms After last query: 25.27 ms Query memory: 0.004 MB Memory before query: 27.675 MB Rows returned: 0
SELECT *
FROM w0v41_jmap_headings
WHERE `linkurl` = 'https://sdat.ir/en/sdat-blog/rootograms,-a-new-way-to-assess-count-models'
| id | select_type | table | partitions | type | possible_keys | key | key_len | ref | rows | filtered | Extra |
|---|
| 1 | SIMPLE | w0v41_jmap_headings | NULL | ALL | NULL | NO INDEX KEY COULD BE USED | NULL | NULL | 1 | 100.00 | Using where |
| Status | Duration |
|---|
| starting | 0.09 ms |
| Executing hook on transaction | 0.01 ms |
| starting | 0.01 ms |
| checking permissions | 0.01 ms |
| Opening tables | 0.10 ms |
| init | 0.01 ms |
| System lock | 0.01 ms |
| optimizing | 0.01 ms |
| statistics | 0.01 ms |
| preparing | 0.02 ms |
| executing | 0.03 ms |
| end | 0.01 ms |
| query end | 0.01 ms |
| waiting for handler commit | 0.01 ms |
| closing tables | 0.01 ms |
| freeing items | 0.07 ms |
| cleaning up | 0.01 ms |
| # | Caller | File and line number |
|---|
| 9 | JDatabaseDriverMysqli->execute() | JROOT/libraries/joomla/database/driver.php:1662 |
| 8 | JDatabaseDriver->loadObject() | JROOT/plugins/system/jmap/jmap.php:819 |
| 7 | plgSystemJMap->onAfterRender() | JROOT/libraries/joomla/event/event.php:70 |
| 6 | JEvent->update() | JROOT/libraries/joomla/event/dispatcher.php:160 |
| 5 | JEventDispatcher->trigger() | JROOT/libraries/src/Application/BaseApplication.php:108 |
| 4 | Joomla\CMS\Application\BaseApplication->triggerEvent() | JROOT/libraries/src/Application/CMSApplication.php:1118 |
| 3 | Joomla\CMS\Application\CMSApplication->render() | JROOT/libraries/src/Application/SiteApplication.php:778 |
| 2 | Joomla\CMS\Application\SiteApplication->render() | JROOT/libraries/src/Application/CMSApplication.php:231 |
| 1 | Joomla\CMS\Application\CMSApplication->execute() | JROOT/index.php:49 |
6 × SELECT *
FROM `w0v41_sppagebuilder`
4 × SELECT enabled
FROM w0v41_extensions
3 × SELECT tag.*
FROM w0v41_k2_tags AS tag JOIN w0v41_k2_tags_xref AS xref
ON tag.id = xref.tagID
3 × SELECT *
FROM w0v41_k2_rating
3 × SELECT *
FROM w0v41_k2_attachments
2 × SELECT `g`.`id`,`g`.`title`
FROM `w0v41_usergroups` AS g
INNER JOIN `w0v41_user_usergroup_map` AS m
ON m.group_id = g.id
2 × SELECT DISTINCT a.id, a.title, a.name, a.checked_out, a.checked_out_time, a.note, a.state, a.access, a.created_time, a.created_user_id, a.ordering, a.language, a.fieldparams, a.params, a.type, a.default_value, a.context, a.group_id, a.label, a.description, a.required,l.title AS language_title, l.image AS language_image,uc.name AS editor,ag.title AS access_level,ua.name AS author_name,g.title AS group_title, g.access as group_access, g.state AS group_state, g.note as group_note
FROM w0v41_fields AS a
LEFT JOIN `w0v41_languages` AS l
ON l.lang_code = a.language
LEFT JOIN w0v41_users AS uc
ON uc.id=a.checked_out
LEFT JOIN w0v41_viewlevels AS ag
ON ag.id = a.access
LEFT JOIN w0v41_users AS ua
ON ua.id = a.created_user_id
LEFT JOIN w0v41_fields_groups AS g
ON g.id = a.group_id
2 × SELECT template
FROM w0v41_template_styles as s
2 × SELECT id, gender, description, image, url, `
group`, plugins
FROM w0v41_k2_users
2 × SELECT *
FROM w0v41_k2_items
2 × SELECT `extension_id`
FROM `w0v41_extensions`
2 × SELECT *
FROM w0v41_extensions
2 × SELECT id, alias, parent
FROM w0v41_k2_categories
2 × SELECT *
FROM `w0v41_users`
1 × SELECT *
FROM w0v41_acymailing_confi
1 × SELECT folder.*
FROM `w0v41_eventgallery_folder` AS folder
LEFT JOIN `w0v41_eventgallery_file` AS file
ON folder.folder = file.folder
and file.published=1
and file.ismainimage=0
1 × SELECT language,id
FROM `w0v41_menu`
1 × SELECT *
FROM w0v41_acymailing_list
1 × SELECT *
FROM `w0v41_acymailing_fields` as a
1 × SELECT *
FROM `w0v41_acymailing_fields
1 × SELECT `c2`.`language`,`c2`.`id`
FROM `w0v41_menu` AS `c`
INNER JOIN `w0v41_associations` AS `a`
ON a.id = c.`id`
AND a.context='com_menus.item'
INNER JOIN `w0v41_associations` AS `a2`
ON `a`.`key` = `a2`.`key`
INNER JOIN `w0v41_menu` AS `c2`
ON a2.id = c2.`id`
1 × SELECT *
FROM w0v41_jmap_metainfo
1 × SELECT *
FROM w0v41_jmap_canonicals
1 × SELECT i.*, c.alias AS categoryalias
FROM w0v41_k2_items AS i
LEFT JOIN w0v41_k2_categories c
ON c.id = i.catid
1 × SELECT `session_id`
FROM `w0v41_session`
1 × SELECT *
FROM w0v41_k2_comments
1 × SELECT DISTINCT a.id, a.title, a.name, a.checked_out, a.checked_out_time, a.note, a.state, a.access, a.created_time, a.created_user_id, a.ordering, a.language, a.fieldparams, a.params, a.type, a.default_value, a.context, a.group_id, a.label, a.description, a.required,l.title AS language_title, l.image AS language_image,uc.name AS editor,ag.title AS access_level,ua.name AS author_name,g.title AS group_title, g.access as group_access, g.state AS group_state, g.note as group_note
FROM w0v41_fields AS a
LEFT JOIN `w0v41_languages` AS l
ON l.lang_code = a.language
LEFT JOIN w0v41_users AS uc
ON uc.id=a.checked_out
LEFT JOIN w0v41_viewlevels AS ag
ON ag.id = a.access
LEFT JOIN w0v41_users AS ua
ON ua.id = a.created_user_id
LEFT JOIN w0v41_fields_groups AS g
ON g.id = a.group_id
LEFT JOIN `w0v41_fields_categories` AS fc
ON fc.field_id = a.id
1 × SELECT id, rules
FROM `w0v41_viewlevels
1 × SELECT *
FROM w0v41_k2_categories
1 × SELECT *
FROM w0v41_k2_items
1 × SELECT id
FROM w0v41_k2_categories
1 × SELECT `id`,`name`,`rules`,`parent_id`
FROM `w0v41_assets`
1 × SELECT id, alias
FROM w0v41_k2_items
1 × SELECT *
FROM w0v41_rsform_confi
1 × SELECT extension, file, type
FROM w0v41_rokcommon_configs
1 × SELECT `name`
FROM `w0v41_extensions`
1 × SELECT b.id
FROM w0v41_usergroups AS a
LEFT JOIN w0v41_usergroups AS b
ON b.lft <= a.lft
AND b.rgt >= a.rgt
1 × SELECT *
FROM w0v41_jmap_headings